# setwd("~/Library/CloudStorage/Box-Box/Teaching/R_2023/local/lec3")
library(haven)Lecture 3
Course Updates
Lecture 1’s link is now available on GitHub
Any Questions? Comments? Suggestions?
Recap from Yesterday
Control structures
- Conditional statements (
if,else,ifelse) - Loops (
for,while,repeat,break,next)
- Conditional statements (
Functions:
- Introduction to built-in functions
- Writing custom functions
- The
applyfamily of functions (lapply,sapply,tapply, etc.)
Library and Packages
Data Manipulation
basetidyversedata.table
Introduction
In this lecture, we will delve into the following topics:
- More in Data Cleaning and Wrangling:
- Revisit the Combining Data sets from yesterday
merge:basejoin:dplyr- Compare with
1:m,m:1,1:1in STATA
- Data Reshaping
data.table
- Data Standardization
- Dates:
- Text:
stringr
- Unique Identifier
- Revisit the Combining Data sets from yesterday
- Basic Data Analysis and Data Visualization in R
- Preliminary Data Checks
- Simple Regression
- Results and Visualization
- A Comprehensive Project in Class
- A review for the last 2.5 days
- Do this in the
rmdfile I provide you. - Your output should be a
htmlreport
For Tomorrow
- Intermediate Econometrics in R Review
- Mathematical Statistics in R
- Distributions
- Simulations
- Intermediate Applied Econometrics
- Testing
- …
- Mathematical Statistics in R
Data Manipulation Again
Merge: Comparison with STATA
In STATA, when merging datasets, you specify the type of merge using:
1:1: One-to-one join: Each observation in the dataset has a unique identifier, and each identifier in one dataset matches to one and only one identifier in the other dataset.1:m: One-to-many join: You start with the “many” dataset. For each unique identifier in the “many” dataset, there’s a corresponding single observation in the “one” dataset. When you perform the merge, each of the multiple observations in the “many” dataset with the same identifier gets matched to a single observation in the “one” dataset.m:1: Many-to-one join: You start with the “one” dataset. For each unique identifier in the “one” dataset, there are multiple corresponding observations in the “many” dataset. When you perform the merge, the single observation in the “one” dataset gets matched to each of the multiple observations in the “many” dataset with the same identifier.
Let’s use examples to understand the link and difference between merging datasets in R and STATA. We will see two cases: 1) all keys can be matched and 2) some keys cannot be matched:
- Demo with
Rand save the temporary data into a.dtafile using thehavenlibrary. - Use
STATAcommands and see if the output matches our expectations.
Every Key Exist in Both Data Set
All three should be the same.
R Demo using base
Since we need to save datasets, we need to define the working directory and libraries we are going to use as usual.
One-to-One (1:1) Merge:
df1 <- data.frame(ID = c(1, 2, 3), Name = c("Alice", "Bob", "Charlie"))
df2 <- data.frame(ID = c(1, 2, 3), Age = c(25, 30, 28))
write_dta(df1, "df1.dta")
write_dta(df2, "df2.dta")
merged_df <- merge(df1, df2, by = "ID")
write_dta(merged_df, "one_to_one_merge.dta")Many-to-One (m:1) Merge:
many_df <- data.frame(ID = c(1, 1, 2, 3), Score = c(85, 90, 88, 92))
one_df <- data.frame(ID = c(1, 2, 3), Name = c("Alice", "Bob", "Charlie"))
write_dta(many_df, "many_df.dta")
write_dta(one_df, "one_df.dta")
merged_df <- merge(many_df, one_df, by = "ID")
write_dta(merged_df, "many_to_one_merge_R.dta")One-to-Many (1:m) Merge:
(Using the same example datasets as for the m:1 merge)
merged_df <- merge(one_df, many_df, by = "ID")
write_dta(merged_df, "one_to_many_merge_R.dta")STATA Demo
We should expect the perfect match.
R Demo with dplyr: left_join and right_join
For this case, the results should be the same. Because both many_df and one_df are having information for all IDs. Let us double-check if we are using left_join, right_join in R.
First, we’ll create two data frames: teachers (the “one” dataset) and classes (the “many” dataset). Then, we’ll perform both m:1 and 1:m merges using both left and right joins, and show the results.
Here’s the R code:
# Load the required library
library(dplyr)
# Create the 'teachers' dataframe (the "one" dataset)
teachers <- data.frame(
TeacherID = c(1, 2),
TeacherName = c("Mr. Smith", "Mrs. Jones")
)
# Create the 'classes' dataframe (the "many" dataset)
classes <- data.frame(
ClassID = c("A", "B", "C"),
ClassName = c("Math", "Science", "English"),
TeacherID = c(1, 1, 2)
)
# m:1 Merge using left join (starting with 'classes' as the base)
m1_merge_left <- left_join(classes, teachers, by = "TeacherID")
# 1:m Merge using left join (starting with 'teachers' as the base)
# This produces the same result as the m:1 merge but potentially with reordered columns
one_m_merge_left <- left_join(teachers, classes, by = "TeacherID")
# m:1 Merge using right join (starting with 'classes' as the base)
m1_merge_right <- right_join(classes, teachers, by = "TeacherID")
# 1:m Merge using right join (starting with 'teachers' as the base)
one_m_merge_right <- right_join(teachers, classes, by = "TeacherID")
# Print results
list(
m1_merge_left = m1_merge_left,
one_m_merge_left = one_m_merge_left,
m1_merge_right = m1_merge_right,
one_m_merge_right = one_m_merge_right
)$m1_merge_left
ClassID ClassName TeacherID TeacherName
1 A Math 1 Mr. Smith
2 B Science 1 Mr. Smith
3 C English 2 Mrs. Jones
$one_m_merge_left
TeacherID TeacherName ClassID ClassName
1 1 Mr. Smith A Math
2 1 Mr. Smith B Science
3 2 Mrs. Jones C English
$m1_merge_right
ClassID ClassName TeacherID TeacherName
1 A Math 1 Mr. Smith
2 B Science 1 Mr. Smith
3 C English 2 Mrs. Jones
$one_m_merge_right
TeacherID TeacherName ClassID ClassName
1 1 Mr. Smith A Math
2 1 Mr. Smith B Science
3 2 Mrs. Jones C EnglishJust like before, everything matched!
Some keys cannot be matched
Now, let us see the behavior of left_join when there’s no match for a record in either the left or the right table, and we want this being demonstrated using both dplyr and base R.
Let’s slightly modify our datasets to have unmatched records:
- In the
teachersdataframe, let’s add a teacher withTeacherID = 3named “Mr. Doe” who doesn’t have any associated class. - In the
classesdataframe, let’s add a class withClassID = "D"named “History” withTeacherID = 4, but there’s no teacher withTeacherID = 4in theteachersdataframe.
Now, let’s perform the left joins using both dplyr and base R:
Using dplyr:
- Perform
left_joinusingclassesas the base. - Perform
left_joinusingteachersas the base.
Using base R:
- Use
mergewithall.x = TRUEusingclassesas the base. - Use
mergewithall.x = TRUEusingteachersas the base.
# Modify the datasets
teachers_modified <- rbind(teachers, data.frame(TeacherID = 3, TeacherName = "Mr. Doe"))
classes_modified <- rbind(classes, data.frame(ClassID = "D", ClassName = "History", TeacherID = 4))
# Using dplyr:
dplyr_classes_base <- left_join(classes_modified, teachers_modified, by = "TeacherID")
dplyr_teachers_base <- left_join(teachers_modified, classes_modified, by = "TeacherID")
# Using base R:
base_classes_base <- merge(classes_modified, teachers_modified, by = "TeacherID", all.x = TRUE)
base_teachers_base <- merge(teachers_modified, classes_modified, by = "TeacherID", all.x = TRUE)
list(
dplyr_classes_base = dplyr_classes_base,
dplyr_teachers_base = dplyr_teachers_base,
base_classes_base = base_classes_base,
base_teachers_base = base_teachers_base
)$dplyr_classes_base
ClassID ClassName TeacherID TeacherName
1 A Math 1 Mr. Smith
2 B Science 1 Mr. Smith
3 C English 2 Mrs. Jones
4 D History 4 <NA>
$dplyr_teachers_base
TeacherID TeacherName ClassID ClassName
1 1 Mr. Smith A Math
2 1 Mr. Smith B Science
3 2 Mrs. Jones C English
4 3 Mr. Doe <NA> <NA>
$base_classes_base
TeacherID ClassID ClassName TeacherName
1 1 A Math Mr. Smith
2 1 B Science Mr. Smith
3 2 C English Mrs. Jones
4 4 D History <NA>
$base_teachers_base
TeacherID TeacherName ClassID ClassName
1 1 Mr. Smith A Math
2 1 Mr. Smith B Science
3 2 Mrs. Jones C English
4 3 Mr. Doe <NA> <NA>NOTES:
In both the dplyr and base R results, we can observe that where there’s no match for a record in the left table, NA values are filled in for columns from the right table. Which should be similar to the m:1 results from STATA’s picture below. (NOT EXACT THE SAME BUT SAME IDEA)
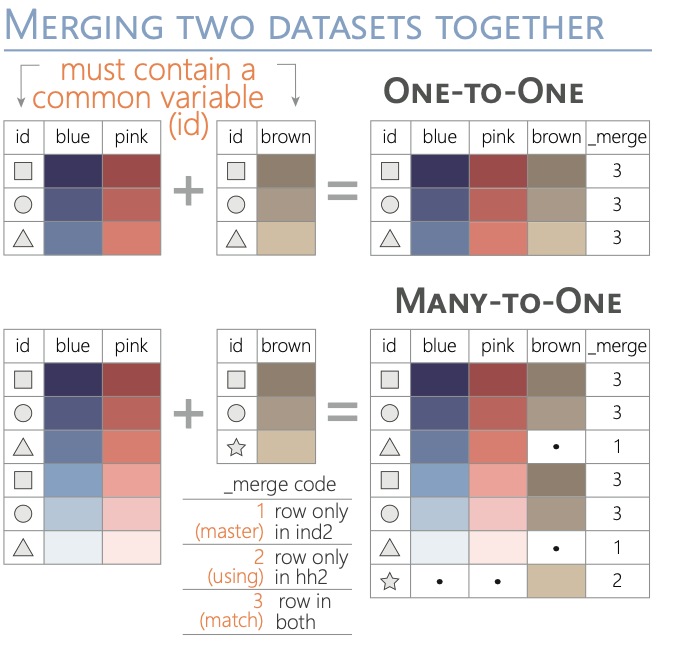
Reshaping
Base R Methods:
reshape Function:
Base R provides the reshape function, which can convert data from wide to long format and vice versa.
Wide to Long:
# Sample data data <- data.frame( ID = 1:3, Time1 = c(5, 6, 7), Time2 = c(8, 9, 10) ) print(data)ID Time1 Time2 1 1 5 8 2 2 6 9 3 3 7 10# Reshaping to long format long_data <- reshape(data, direction = "long", varying = list(c("Time1", "Time2")), v.names = "Value", idvar = "ID", timevar = "Time") print(long_data)ID Time Value 1.1 1 1 5 2.1 2 1 6 3.1 3 1 7 1.2 1 2 8 2.2 2 2 9 3.2 3 2 10Long to Wide:
# Reshaping to wide format wide_data <- reshape(long_data, direction = "wide", v.names = "Value", idvar = "ID", timevar = "Time") print(wide_data)ID Value.1 Value.2 1.1 1 5 8 2.1 2 6 9 3.1 3 7 10
Using data.table:
The data.table library offers an efficient and flexible approach to data reshaping, especially for large datasets.
Melting (Wide to Long):
library(data.table)
# Convert data frame to data table
DT <- as.data.table(data)
# Melt to long format
melted_data <- melt(DT, id.vars = "ID", measure.vars = c("Time1", "Time2"),
variable.name = "Time", value.name = "Value")
print(melted_data) ID Time Value
<int> <fctr> <num>
1: 1 Time1 5
2: 2 Time1 6
3: 3 Time1 7
4: 1 Time2 8
5: 2 Time2 9
6: 3 Time2 10Casting (Long to Wide):
# Cast to wide format
casted_data <- dcast(melted_data, ID ~ Time, value.var = "Value")
print(casted_data)Key: <ID>
ID Time1 Time2
<int> <num> <num>
1: 1 5 8
2: 2 6 9
3: 3 7 10Remember, the choice between base R and data.table methods often depends on your specific needs. data.table is especially powerful for large datasets due to its efficiency, while base R can be simpler for basic reshaping tasks or for those who are more familiar with its syntax.
Dates
- Date data can be tricky due to various formats and conventions across the world.
- Proper handling is essential for chronological analysis, time series forecasting, and event tracking.
Sys.Date(): Returns the current date.as.Date(): Converts a character string into a Date object.format(): Formats a Date object into a desired character representation.
Examples
# Current date
current_date <- Sys.Date()
print(current_date)[1] "2025-09-17"# Convert a string to a date
date_str <- "2023-08-16"
converted_date <- as.Date(date_str)
print(converted_date)[1] "2023-08-16"# Format a date
formatted_date <- format(current_date, format="%B %d, %Y")
print(formatted_date)[1] "September 17, 2025"# Define starting and ending dates
start_date <- as.Date("2023-01-01")
end_date <- as.Date("2023-01-10")
# Create a sequence of dates
date_seq <- seq(start_date, end_date, by="days")
# Convert the sequence to a data frame
date_df <- data.frame(Date = date_seq)
# Print the data frame
print(date_df) Date
1 2023-01-01
2 2023-01-02
3 2023-01-03
4 2023-01-04
5 2023-01-05
6 2023-01-06
7 2023-01-07
8 2023-01-08
9 2023-01-09
10 2023-01-10Strings
Text data often contains noise in the form of special characters, inconsistencies in formatting, and more. Properly cleaning and manipulating such data is crucial.
- The
stringrpackage in R offers a host of functions that can aid in this. gsubinbaseR can do a lot similar functions, I will leave that to you for the future study. Some examples have been provided in the Appendix Section.
Removing Special Characters
- Special characters can be noise in some analyses.
- Removing them can simplify text and aid in other text processing tasks.
str_replace_all(): Replaces all instances of a pattern in a string.
Examples
library(stringr)
text <- "Hello, world! This is a test. #Test123"
cleaned_text <- str_replace_all(text, "[^[:alnum:][:space:]]", "")
print(cleaned_text)[1] "Hello world This is a test Test123"String Matching
- Useful to detect if a string contains certain patterns or characters.
str_detect(): Detects the presence or absence of a pattern in a string.str_which(): Returns the indices of strings that match a pattern.str_match(): Extract matched groups from a string based on a pattern.
Examples
# Detect if a string contains "world"
text <- c("Hello world", "Hello R", "R is a world of statistics")
print(str_detect(text, "world"))[1] TRUE FALSE TRUE# Get indices of strings that contain "R"
print(str_which(text, "R"))[1] 2 3# Extract matched groups
pattern <- "(\\d{4})-(\\d{2})-(\\d{2})"
date_str <- "Today's date is 2023-08-16."
print(str_match(date_str, pattern)) [,1] [,2] [,3] [,4]
[1,] "2023-08-16" "2023" "08" "16"Case Conversion
- To ensure uniformity in text data.
str_to_upper(): Converts strings to upper case.str_to_lower(): Converts strings to lower case.str_to_title(): Converts strings to title case.
Examples
text <- "Hello, World!"
print(str_to_upper(text))[1] "HELLO, WORLD!"print(str_to_lower(text))[1] "hello, world!"print(str_to_title(text))[1] "Hello, World!"Splitting Strings
- To break a string into parts based on a delimiter.
str_split(): Splits a string into parts.
Examples
text <- "apple,banana,grape"
print(str_split(text, ","))[[1]]
[1] "apple" "banana" "grape" Other stringr functions in R with examples
str_length(): Computes the length of a string.str_c(): Concatenates strings.str_sub(): Extracts or replaces substrings.
library(stringr)
# String length
print(str_length("Hello, world!"))[1] 13# Concatenate
print(str_c("Hello", "world", sep=", "))[1] "Hello, world"# Substring
print(str_sub("Hello, world!", 1, 5))[1] "Hello"Unique Identifiers in R
- A unique identifier (UID) is an identifier that ensures distinctness among all other items.
- UIDs are crucial for indexing, referencing, and joining datasets without confusion.
Functions to Identify Unique and Duplicate Values
unique(): Returns a vector of unique values.
vec <- c(1, 2, 2, 3, 4, 4, 4, 5)
unique_vals <- unique(vec)
print(unique_vals)[1] 1 2 3 4 5duplicated(): Returns a logical vector indicating whether an element is a duplicate.
vec <- c(1, 2, 2, 3, 4, 4, 4, 5)
dupes <- duplicated(vec)
print(dupes)[1] FALSE FALSE TRUE FALSE FALSE TRUE TRUE FALSEdistinct() from the dplyr package: Used to remove duplicate rows from a data frame or tibble.
library(dplyr)
df <- data.frame(name = c("Alice", "Bob", "Alice", "Charlie"), age = c(25, 30, 25, 35))
distinct_df <- distinct(df)
print(distinct_df) name age
1 Alice 25
2 Bob 30
3 Charlie 35Generating Unique Identifiers
seq_along(): Generate a sequence along an object’s length.
vec <- c("apple", "banana", "cherry")
uids <- seq_along(vec)
print(uids)[1] 1 2 3make.unique(): Generates unique strings by appending numbers.
vec <- c("apple", "apple", "banana")
unique_vec <- make.unique(vec)
print(unique_vec)[1] "apple" "apple.1" "banana" Checking for Unique Identifiers with isid()
The isid() function checks if a given set of variables uniquely identifies the observations in a dataset. Just like the one we are using in STATA.
- You need to install
eeptoolspackage for this.
library(eeptools)
df <- data.frame(id = c(1, 2, 3, 1), value = c(10, 20, 30, 40))
# Check if 'id' uniquely identifies the data
isid(df, "id", verbose = TRUE)Are variables a unique ID?
[1] FALSE
Variables define this many unique rows:
[1] 3
There are this many total rows in the data:
[1] 4In this example, the isid() function would return FALSE because the ‘id’ variable does not uniquely identify each row in the dataset.
Considerations
- When working with large datasets or datasets that will be merged, it’s essential to ensure that the data’s unique identifiers remain consistent.
- Always check for the uniqueness of identifiers, especially before operations like joining or merging datasets, to prevent unintended duplications or omissions.
Basic Data Analysis and Data Visulizations in R
Preliminary Checks
Before diving into any data analysis, it’s essential to understand and clean your data. This involves checking for outliers, handling missing values, and visualizing data distributions.
For our demonstration, we’ll use the
mtcarsdataset.data(mtcars)
Checking for Outliers
Outliers can significantly affect regression results. A simple way to check for outliers is by using boxplots.
boxplot(mtcars$mpg, main="Boxplot of MPG", ylab="Miles Per Gallon")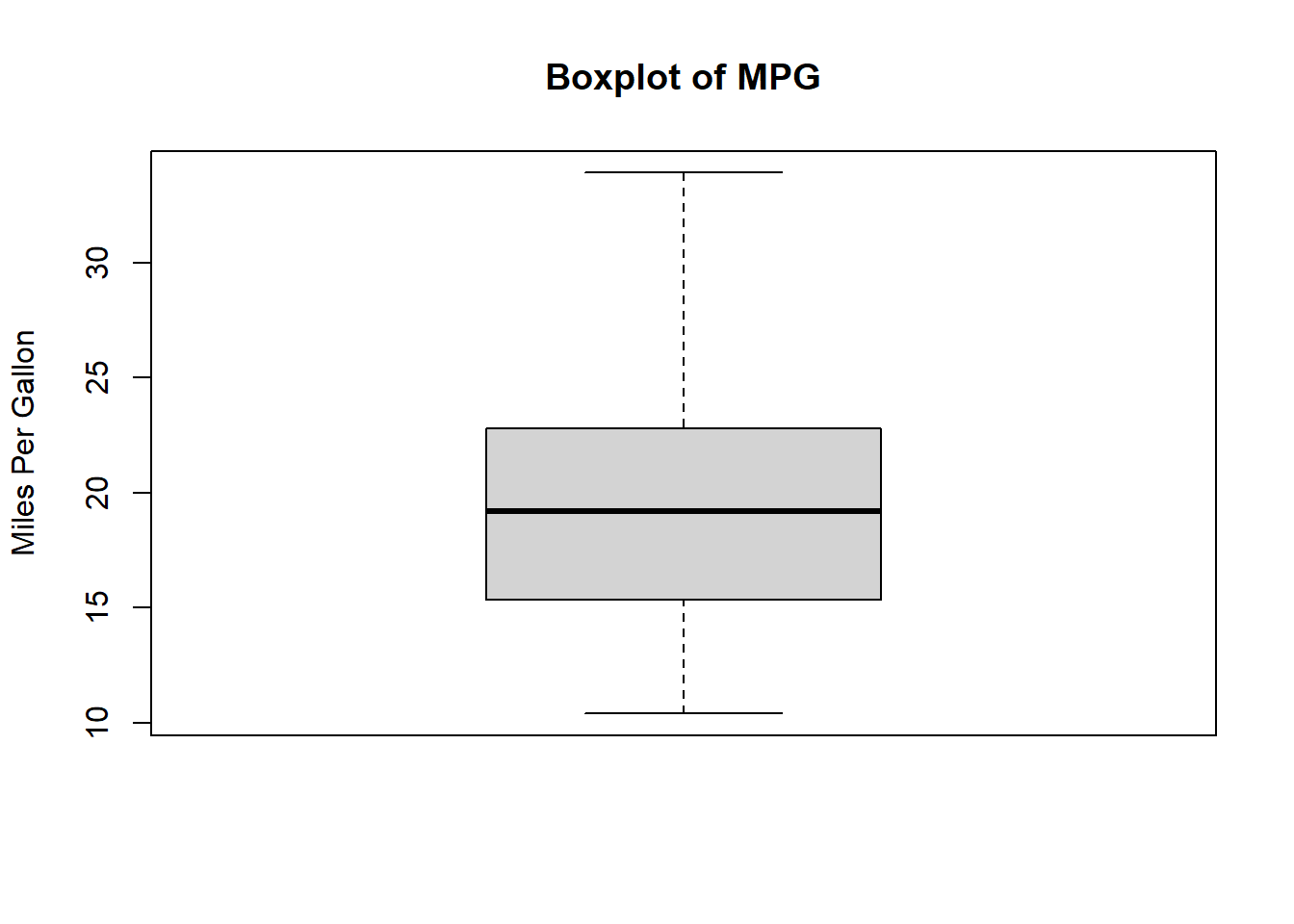
Points outside the “whiskers” of the boxplot could be potential outliers.
Handling Missing Values
Data often comes with missing values, and it’s crucial to handle them appropriately.
Identifying Missing Values
Use
is.na()to identify missing values:
missing_vals <- is.na(mtcars$mpg)
sum(missing_vals)[1] 0- Handling Strategies
- Remove rows with missing values:
na.omit() - Impute missing values using mean, median, or a specific strategy.
- Remove rows with missing values:
Data Visualization
Visualizing your data can help in understanding distributions, relationships, and potential issues.
- Histogram: Understand the distribution of a variable.
hist(mtcars$mpg, main="Histogram of MPG", xlab="Miles Per Gallon", col="lightblue")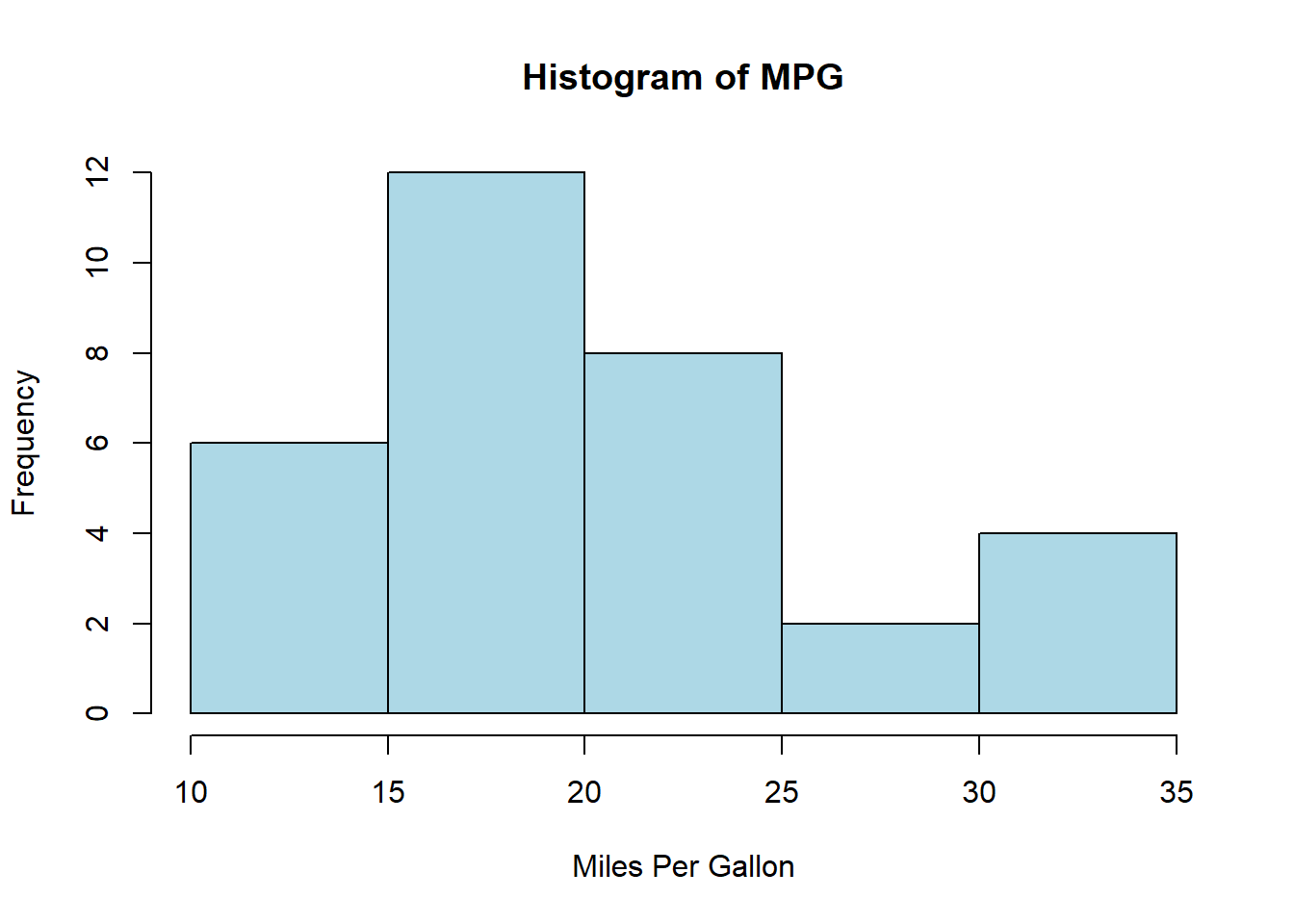
- Correlation Plot: Understand relationships between variables.
pairs(mtcars[, 1:4], main="Scatterplot Matrix")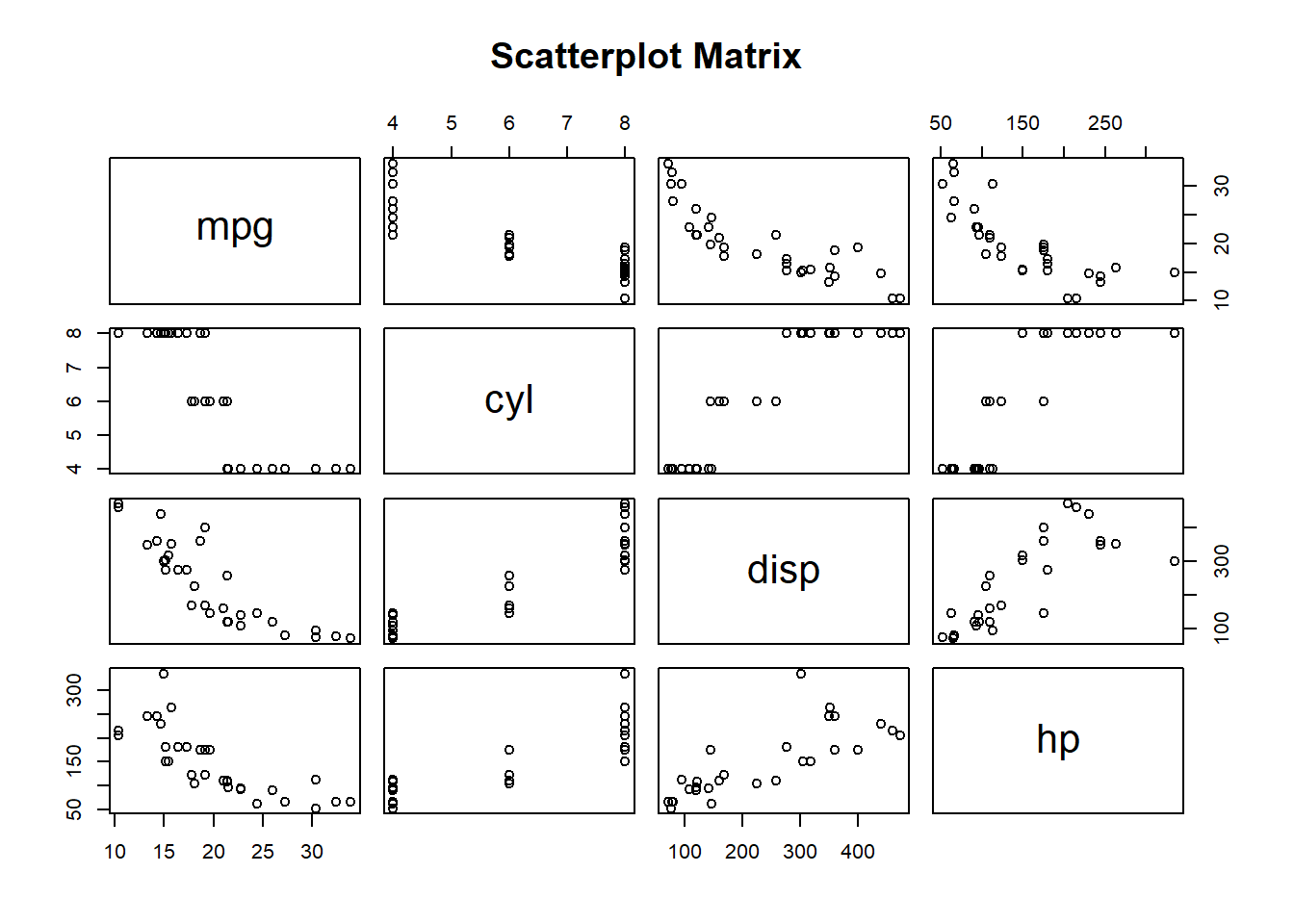
- Density Plot: Another way to check the distribution.
ggplot(mtcars, aes(x=mpg)) +
geom_density(fill="blue", alpha=0.5) +
labs(title="Density Plot of MPG", x="Miles Per Gallon")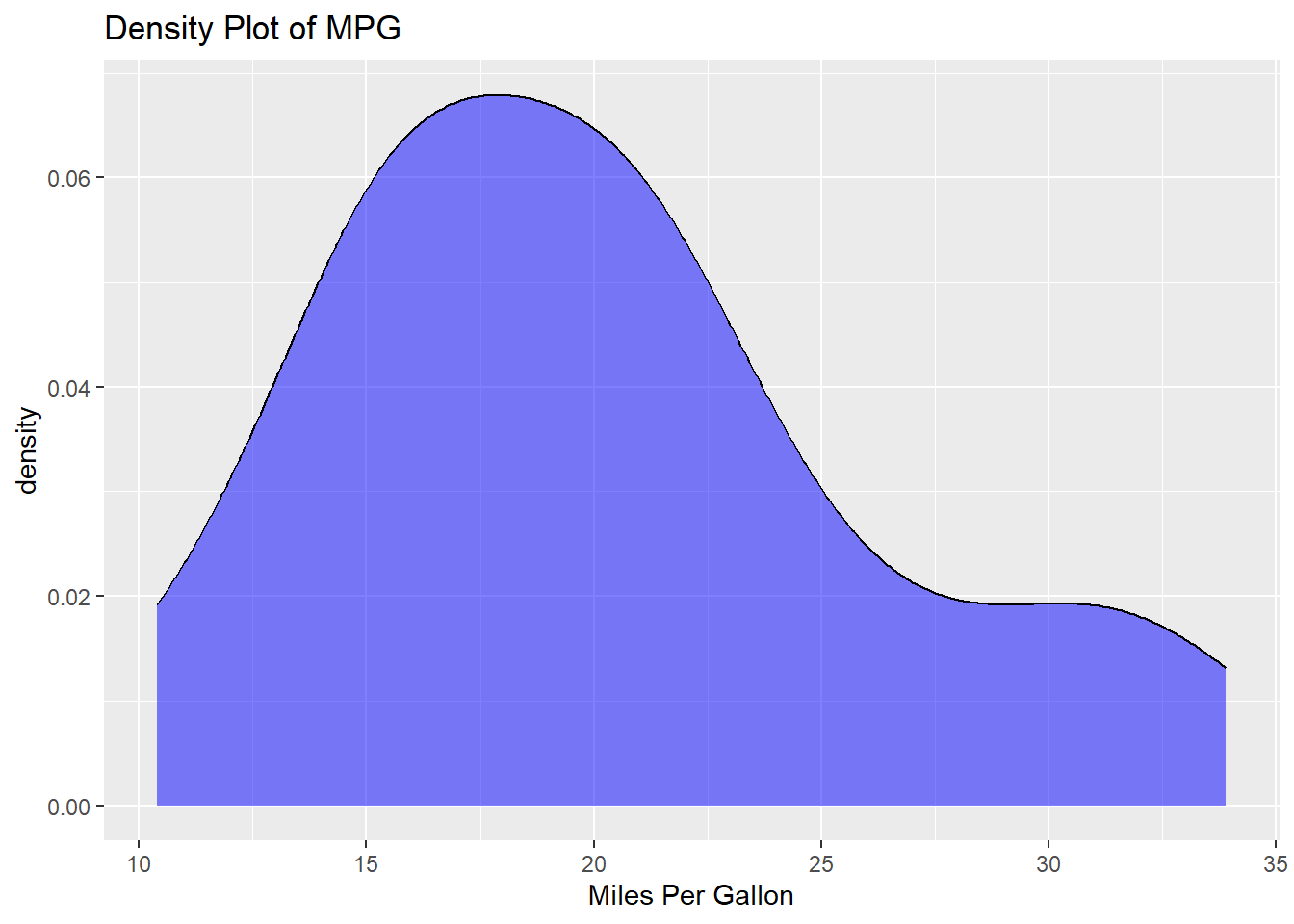
By conducting these preliminary checks, you ensure that your data is ready for deeper analysis, and any insights or results derived are more likely to be reliable.
Data Analysis and Data Visulization (mtcars example)
Make sure you’ve installed the packages:
tidyverse,lmtestDone the first-round preliminary data check.
Loading the Libraries
library(tidyverse)
library(lmtest)Running a Simple Regression in R
A regression allows us to understand relationships between variables. The simplest form is the linear regression, represented as:
\[ Y = \beta_0 + \beta_1 X + \epsilon \]
Where: - \(Y\) is the dependent variable. - \(X\) is the independent variable. - \(\beta_0\) is the intercept. - \(\beta_1\) is the slope. - \(\epsilon\) is the error term.
model <- lm(mpg ~ wt, data = mtcars)
summary(model)
Call:
lm(formula = mpg ~ wt, data = mtcars)
Residuals:
Min 1Q Median 3Q Max
-4.5432 -2.3647 -0.1252 1.4096 6.8727
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 37.2851 1.8776 19.858 < 2e-16 ***
wt -5.3445 0.5591 -9.559 1.29e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.046 on 30 degrees of freedom
Multiple R-squared: 0.7528, Adjusted R-squared: 0.7446
F-statistic: 91.38 on 1 and 30 DF, p-value: 1.294e-10The summary() function provides a detailed summary of the regression results. Here, we are trying to predict mpg (miles per gallon) using the weight (wt) of the car, using the mtcars dataset.
Accessing Regression Results
We can save the summary of the model first:
model_summary <- summary(model)And we can see that it is a list! This is something we’ve already learned.
From this summary object, you can extract:
coefficients: A matrix where each row represents a predictor (including the intercept) and columns provide details like estimate, standard error, t-value, and p-value.sigma: The residual standard error.r.squared: The R-squared value.adj.r.squared: The adjusted R-squared value.fstatistic: The F-statistic value and its degrees of freedom.
- Access the Coefficients
# Access using summary list
coeff_matrix <- model_summary$coefficients
print(coeff_matrix) Estimate Std. Error t value Pr(>|t|)
(Intercept) 37.285126 1.877627 19.857575 8.241799e-19
wt -5.344472 0.559101 -9.559044 1.293959e-10# Directly access using `coefficients` functions for regression objects
betas <- coefficients(model) # this will give you only the betas
print(betas)(Intercept) wt
37.285126 -5.344472 - Access the Standard Errors
The standard errors of the coefficients can be extracted from the model’s summary object.
standard_errors <- coeff_matrix[, "Std. Error"]
print(standard_errors)(Intercept) wt
1.877627 0.559101 - Access the t-statistics
t_values <- summary(model)$coefficients[, "t value"]
print(t_values)(Intercept) wt
19.857575 -9.559044 - Access the p-values
p_values <- summary(model)$coefficients[, "Pr(>|t|)"]
print(p_values) (Intercept) wt
8.241799e-19 1.293959e-10 4. Show Basic Regression Figures
Scatter Plot with Regression Line
Visualizing the relationship between the independent and dependent variables can be very insightful.
ggplot(mtcars, aes(x=wt, y=mpg)) +
geom_point() +
geom_smooth(method="lm", col="red") +
labs(title="Relationship between Car Weight and MPG", x="Weight", y="Miles Per Gallon")`geom_smooth()` using formula = 'y ~ x'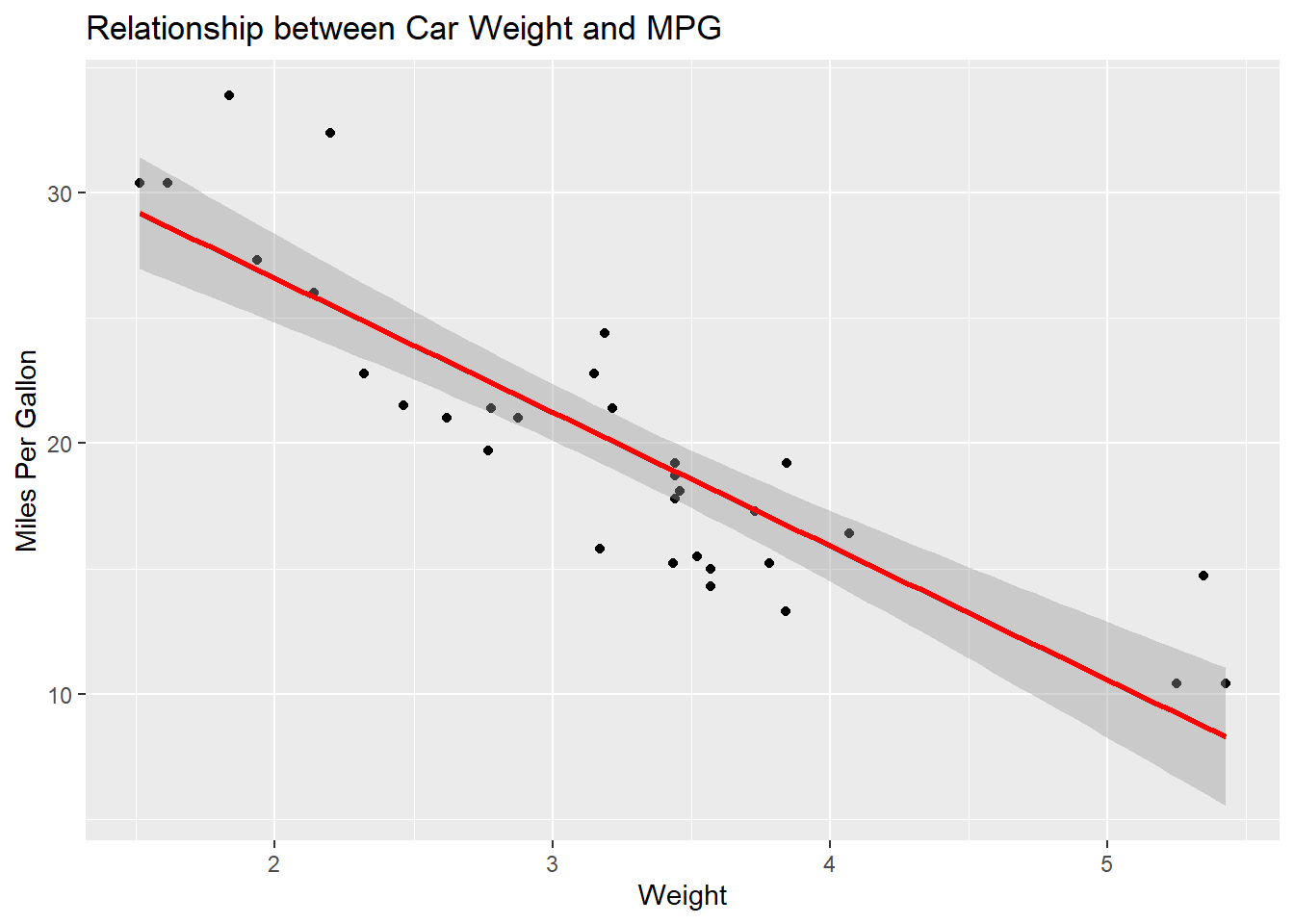
Residuals vs. Fitted Values
Checking the residuals can help diagnose potential issues with the model.
residuals <- resid(model)
fitted_values <- fitted(model)
ggplot() +
geom_point(aes(x=fitted_values, y=residuals)) +
labs(title="Residuals vs. Fitted Values", x="Fitted Values", y="Residuals")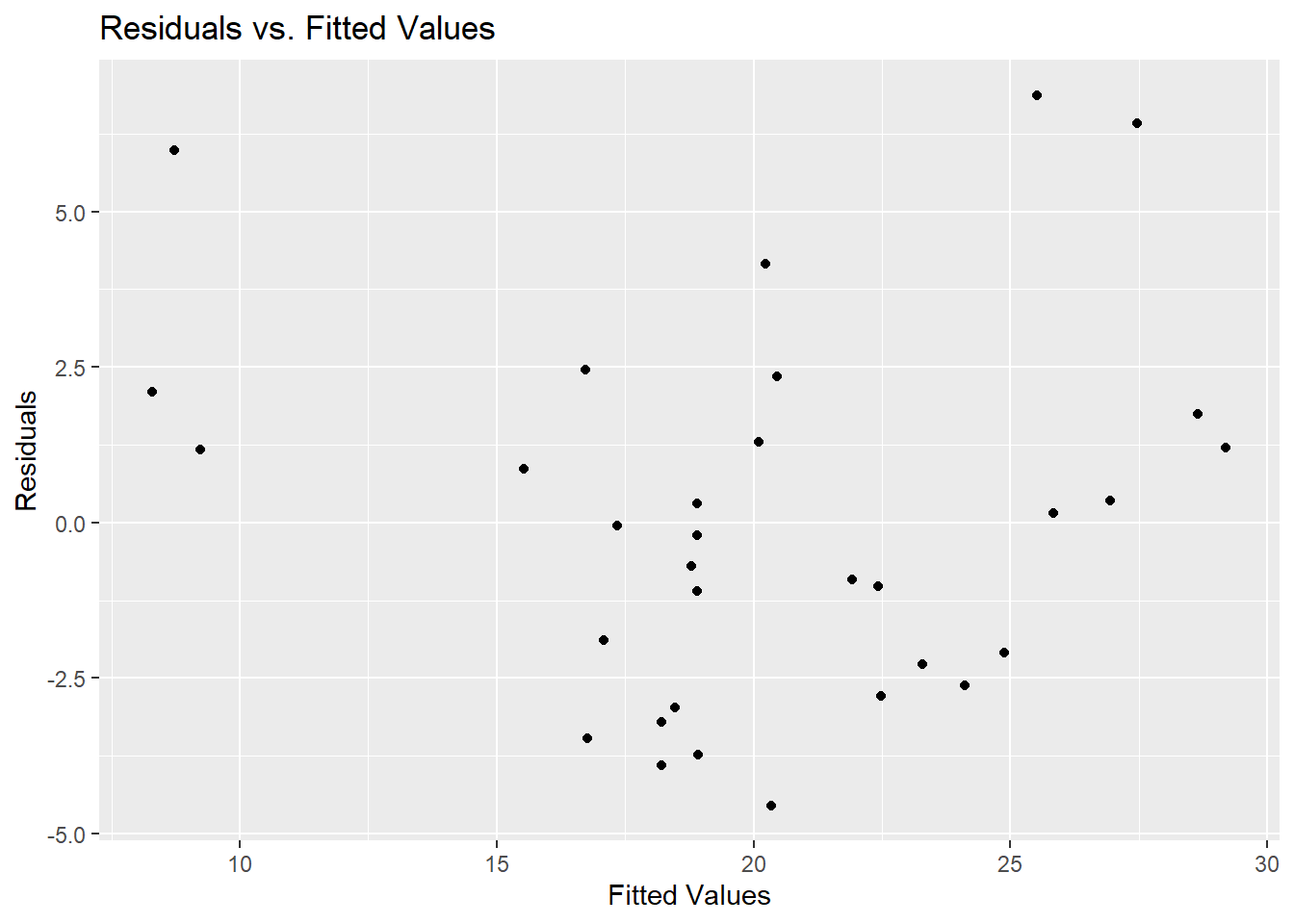
Comprehensive Reivew Project
Introduction
This comprehensive project is designed to review the concepts you’ve learned throughout the course. You’ll apply techniques from data cleaning to advanced data analysis. We’ll work with both built-in datasets and an external dataset.
Datasets
- Built-in Dataset:
mtcars - External Dataset: “client_data.csv”
Workflows
Create a workflow for this project in the following steps:
Under lec3 folder, create a sub folder lec3_proj
Under lec3_proj folder, create two sub folders: code and data
Under data folder, create three sub folders: raw, temp, and cleaned
Your directories should look like this:
- lec3–>
- lec3_proj–>
- code–>
lec3_proj.Rmd
- data–>
- raw
- temp
- cleaned
- code–>
- lec3_proj–>
Tasks
- Preparation
- Download the
lec3_proj.Rmdfrom GitHub into your code folder - It is already setup for you with the questions.
- Code in the code chunk.
- Set up your working directory to the folder
~/lec3/lec3_proj/code/
- Data Exploration and Cleaning:
- Generate an external dataset “client_data.csv” with the following code
set.seed(123)
# Number of clients
n_clients <- 200
# 1. Generate 'name' column
client_data <- data.frame(name = paste0("Client_", seq(1, n_clients)))
# 2. Generate 'car_bought' column. Initially, just a random sample
client_data$car_bought <- sample(rownames(mtcars), n_clients, replace = TRUE)
# 3. Generate 'date_purchased' column
client_data$date_purchased <- sample(seq(as.Date('2015/01/01'), as.Date('2022/01/01'), by="day"), n_clients, replace=TRUE)
# 4. Generate 'income' column
client_data$income <- runif(n_clients, min=30000, max=150000)
# 5. Ensure richer clients are more likely to buy cars with more horsepower
# Sort mtcars by horsepower
sorted_cars <- rownames(mtcars[order(mtcars$hp), ])
# Divide clients into groups and assign cars based on sorted horsepower
split_rows <- ceiling(n_clients / length(sorted_cars))
client_data <- client_data[order(-client_data$income), ] # sort by income
client_data$car_bought <- rep(sorted_cars, times = split_rows)[1:n_clients]
# Show the transformed client_data
head(client_data)
# Write the data to a CSV
write.csv(client_data, "../data/raw/client_data.csv", row.names = FALSE)- Load the built-in dataset
mtcars. - Create a column namedID, which is the same as the rownames. - Identify missing values using
sum, andis.na()function. - Visualize and treat any outliers in the
mpgcolumn of themtcarsdataset. - Hint: usingboxplot()function, and use?boxplot()to learn the syntax.
- Combining Datasets:
- Merge the datasets using
car_ID, andcar_bought. - Use both
mergefrombaseandjoinfromdplyrto combine. Discuss the differences. - Note: the merged dataset should be named as:
merged_data_basemerged_data_dplyr
- Merge the datasets using
- Data Reshaping:
- Use
data.tableto reshape the merged data set. - Save the reshaped data set into the
tempfolder. - Create a summary table calculating the mean
mpgfor each unique value inCar_ID.
- Use
- Data Standardization:
- Standardize any date columns in
client_data.csv. - Purchased Date:
as.Date() - Client’s name: get rid of
Client_usingstr_replace()
- Standardize any date columns in
- Unique Identifier Check:
- Check if the
nameis a unique identifier using: isid()length(unique(DATA$UID))==nrow(DATA)
- Check if the
5.5. Save and Use Cleaned Data to Proceed - Save the “merged_data_base” into the cleaned folder, named as: merged_data_base_cleaned.csv
Regression Analysis:
- Conducting the Regression:
- Using the merged dataset, run a regression predicting
mpgfrom another continuous variable.mpg ~ wt
- Visualize the relationship between the chosen predictor and
mpgusing a scatter plot.plot(merged_data_base$wt, merged_data_base$mpg, ...)abline()
- Using the merged dataset, run a regression predicting
- Matrix Operations with Coefficients:
- Extract the coefficients from the regression model and store them in a matrix.
- new matrix:
matrix() - extract coefficients:
coefficients(model)
- new matrix:
- Create a 2x2 identity matrix.
- Use this:
identity_matrix <- diag(2)
- Use this:
- Use matrix multiplication (using
%*%) to multiply the identity matrix by the coefficients matrix. The result will be the coefficients themselves.
- Extract the coefficients from the regression model and store them in a matrix.
- Loops for Analysis (HARD: Don’t Do it for Now):
- Write a
forloop that iterates over the column names of the merged dataset (excludingmpg). In each iteration, run a regression usingmpgas the dependent variable and the current column as the independent variable. Store each coefficient in a vector. - After the loop, visualize the coefficients using a bar plot to see the impact of each variable on
mpg.
- Write a
- Interpretation:
- Discuss the results. Which variables have the most substantial impact on
mpg? Are the results consistent with expectations?
- Discuss the results. Which variables have the most substantial impact on
- Conducting the Regression:
Control Structures and Custom Functions:
- Use a
forloop to calculate the mean of each numeric column in the merged dataset. (HARD: Don’t Do it now). Hints:- To calculate the mean of each numeric column in a dataset:
- Initialization: Start by creating an empty numeric vector to store the mean values.
- Looping: Use a for loop to iterate over each column in the dataset.
- Conditional Check: Within the loop, check if the current column is numeric. You can use the is.numeric() function for this.
- Calculation: If the column is numeric, calculate its mean using the mean() function. Make sure to handle any missing values. Append this mean value to your storage vector.
- Naming: After the loop completes, name each element in your storage vector with the corresponding column name.
- Output: Finally, print the named vector to display the mean values.
- Write a custom function that uses
ifelseto categorizempginto “Low”, “Medium”, “High”. Apply this function to the dataset.- Hint:
ifelse(..., ifelse(,...))
- Hint:
- Use a
whileloop to find the first row in the merged dataset wherempgis above a certain threshold (e.g., 25).
- The
applyFamily:- Use
lapplyto calculate the range of each numeric column in the merged dataset. (HARD, don’t do it now) - hint:
range_function <- function(x) c(min=min(x, na.rm=TRUE), max=max(x, na.rm=TRUE)) - Use
sapplyto get the type of each column in the dataset. - hint:
sapply(data, class)
- Use
- Advanced Data Manipulation:
- Use
tidyversefunctions to filter rows, select columns, and arrange the dataset. - filter: mpg > 20
- select: mpg, wt, gear
- arrange: desc(wt)
- Use
data.tableto efficiently modify the dataset in place.
- Use
Challenging Question
- Integrative Analysis:
- Create a new column in the merged dataset that calculates the age of the car (assumes everyone bought a new car on the purchase_date). Use any necessary libraries/packages.
- Write a custom function that, given a column name, returns a list containing the mean, median, and standard deviation. Apply this to multiple columns using the
sapplyfunction. - Use control structures to find the average
mpgfor cars that are above and below the median weight. Compare the results and provide an interpretation.
Solution
You can find the solution here.
Appendix
gsub
The gsub() function is part of base R, and it’s a powerful tool for replacing patterns in strings. gsub() stands for “global substitution”. It searches for all matches of a pattern in a string and replaces them with a specified replacement string.
Syntax
gsub(pattern, replacement, x, ignore.case = FALSE, perl = FALSE, fixed = FALSE, useBytes = FALSE)pattern: The pattern to search for.replacement: The string to replace the pattern with.x: The input string.ignore.case: Should the match be case-insensitive?perl: Should Perl-compatible regex be used?fixed: If TRUE, pattern is a string to be matched as is (turns off special characters).useBytes: Should bytes be used for matching (relevant for non-ASCII strings)?
Examples
- Basic Substitution Replacing “cat” with “dog”:
text <- "The cat sat on the mat."
new_text <- gsub("cat", "dog", text)
print(new_text)[1] "The dog sat on the mat."- Removing Special Characters Here’s an example using
gsub()to remove special characters:
text <- "Hello, world! This is a test. #Test123"
cleaned_text <- gsub("[^[:alnum:][:space:]]", "", text)
print(cleaned_text)[1] "Hello world This is a test Test123"- Case-Insensitive Replacement Replacing “world” with “R”, ignoring case:
text <- "Hello World"
new_text <- gsub("world", "R", text, ignore.case = TRUE)
print(new_text)[1] "Hello R"- Replace Multiple Spaces with Single Space
text <- "This has multiple spaces."
new_text <- gsub("\\s+", " ", text)
print(new_text)[1] "This has multiple spaces."gsub() is one of the primary string manipulation functions in base R and is often used in scenarios where you don’t want to or can’t rely on external packages.
Reference
Dr. Qingxiao Li’s notes for R-Review 2020
Rodrigo Franco’s notes for R-Review 2021